Update! If you missed the online tutorial, you can now follow it at your own pace on Youtube:
Session 1: https://youtu.be/xg-LU81UeG0
Session 2: https://youtu.be/bsA0KyKWZXY
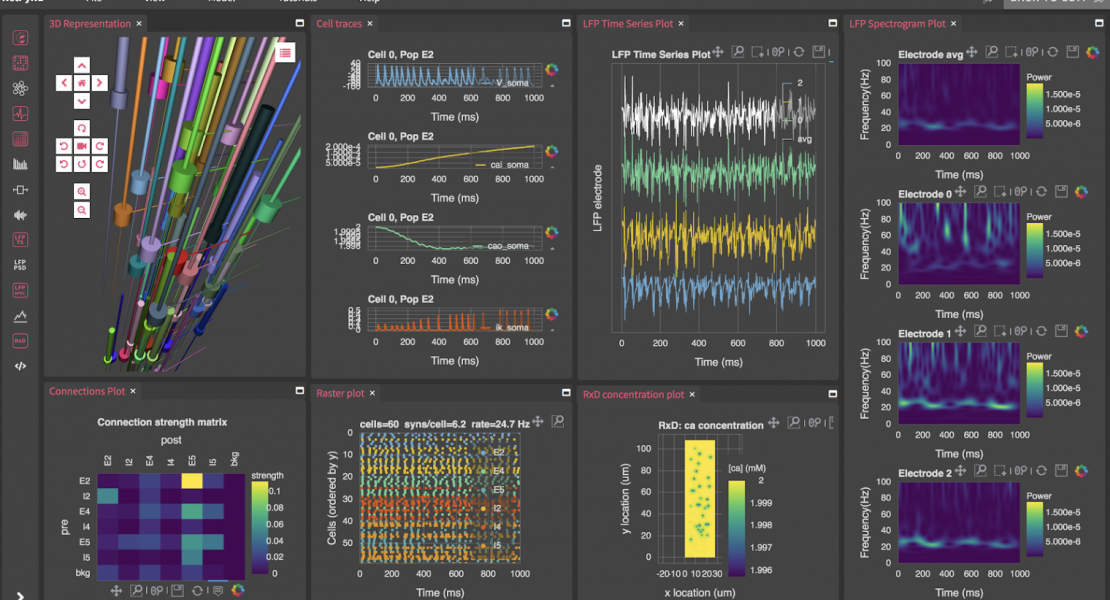
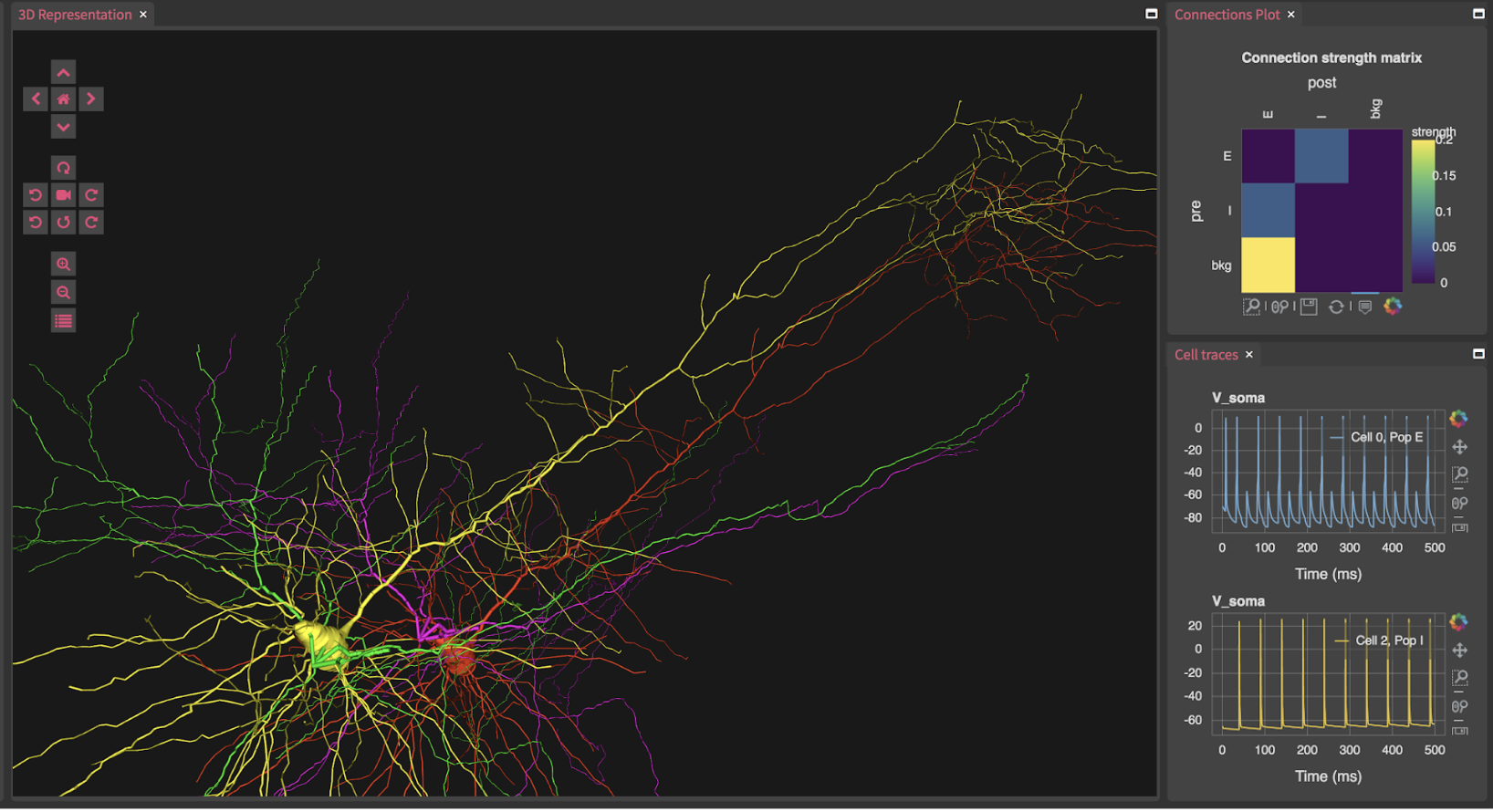
Our online hands-on tutorial provides an introduction to building brain circuit models at multiple scales (molecular, cellular, network, system, behavior) using the NIH-funded NetPyNE [1] and NEURON [2] tools. For more details on the software tool see our eLife publication and tool website.
When: Wednesday June 29, 2022, from 13.00 to 19.00 (UTC); 09.00 to 15.00 (ET)
Where: Online: Zoom link (https://zoom.us/j/92430215843)
Registration: Free but required: Registration form.
Tentative Schedule (ET times):
09.00-09.30 (30min): Introduction to NetPyNE
09.30-12.00 (2h30min): Graphical Web App Tutorials
- Overview Web App and OSB environment
- Simple network
- Network with morphologically detailed neurons
- Multiscale network: from reaction-diffusion to LFP/EEG
- Automated network parameter exploration
12.00-12.30 (30min): Break
12.30-15.00 (2h30min): Jupyter Notebook Programmatic Tutorials
- OSB JupyterLab environment
- Network oscillations tutorial
- NetPyNE data structures
- Importing cell models from Python, HOC, SWC
- Analysis and plotting (including LFP, CSD and EEG)
- Automated network parameter exploration
Requirements: Basic familiarity with Python is recommended. If you’re new to Python programming, there are many excellent tutorials online. There are Python lectures with exercises and solutions taught by one of us in a summer course last year under “Monday” at https://ycmi.github.io/summer-course-2020/.
No prior knowledge of NEURON or NetPyNE is required, but we strongly recommend taking the INCF/CNS Online NEURON course on June 28, 2022. The tutorial will use these tools on the cloud via the Open Source Brain v2.0 platform, so no software installation is necessary.
Download the tutorial slides here: bit.ly/netpyne-online-tutorial-2022
Organizers: The course is organized by the Dura-Bernal Lab and the Neurosim lab at the State University of New York (SUNY) Downstate Health Sciences University. The course is funded by the National Institutes of Health (NIH) National Institutes of Biomedical Imaging and Bioengineering (NIBIB).
The course is part of a series of free online satellite tutorials at CNS*2022 conference organized by The International Neuroinformatics Coordinating Facility (INCF) and the Organization for Computational Neurosciences (OCNS) Software Working Group (WG). Please see this page for more details.
References and background reading:
[1] Dura-Bernal S, Suter B, Gleeson P, Cantarelli M, Quintana A, Rodriguez F, Kedziora DJ, Chadderdon GL, Kerr CC, Neymotin SA, McDougal R, Hines M, Shepherd GMG, Lytton WW. (2019) NetPyNE: a tool for data-driven multiscale modeling of brain circuits. eLife 8:e44494
[2] Omar Awile, Pramod Kumbhar, Nicolas Cornu, Salvador Dura-Bernal, James Gonzalo King, Olli Lupton, Ioannis Magkanaris, Robert A. McDougal, Adam J.H. Newton, Fernando Pereira, Alexandru Săvulescu, Nicholas T. Carnevale, William W. Lytton, Michael L. Hines, Felix Schürmann. Modernizing the NEURON Simulator for Sustainability, Portability, and Performance. Front. Neuroinform., 2022, https://doi.org/10.3389/fninf.2022.884046
[3] Gleeson P, Cantarelli M, Quintana A, Earnsah M, Piasini E, Birgiolas J, Cannon RC, Cayco- Gajic A, Crook S, Davison AP, Dura-Bernal S, Ecker A, Hines ML, Idili G, Lanore F, Larson S, Lytton WW, Majumdar A, McDougal RA, Sivagnanam S, Solinas S, Stanislovas R, van Albada S, van Geit W, Silver RA. (2019) Open Source Brain: a collaborative resource for visualizing, analyzing, simulating and developing standardized models of neurons and circuits. Neuron, 10.1016/j.neuron.2019.05.019.